By Benjamin Louis | 3 Apr 2019 |
What we need, what we want
Besides specific packages presented later, this work is all about manipulating data and of course, tidyverse packages are our friends here. I used dplyr for general data manipulation, purrr for fonctionnal programming and stringr for strings manipulation.
library(dplyr)
library(purrr)
library(stringr)As I cannot publicly share the orginal questionnaire, I made a dummy one from my Google account by mixing proposed templates. The PDF file could be easily downloaded from the Google Form account. For HTML, I first wanted to directly reach the page from the editable version of the questionnaire, using the rvest package, but I never succeeded as it seems Google double authentification makes things difficult. If somebody with better skills for this kind of problem have a solution, I’ll take it ! Instead of performing classic webscraping, I saved the HTML page from my browser and worked with the file locally. All materials are available here.
From these files, the goal is to get a final data.frame with three columns :
questions: the questions’ titlestypes: the types of questions which can take 4 values :uniquefor multiple choices questions with a unique answer,multiplefor multiple choices questions with possibility of several answers,freefor questions with free answer andgridfor mutliple choices grid questions.choices: a list-column with each row being a vector of answer choices foruniqueandmultiplequestions, a named vector forgridquestions (names coresponding to row1, row2, …, column1, …) and vector of length 0 forfreequestions
A try with pdftools
To work with the PDF, I used the rOpenSci’s Jeroen Ooms’ pdftools package. If you don’t know this package, I really encourage you to take a look at it as well as Maëlle Salmon’s post about Trump’s PDF schedule. For Ubuntu users, installation of pdftools can be challenging as the required latest version of Poppler is not available on Linux. I found this thread really useful to deal with the problem (tested on Ubuntu 18.04).
Importing data from a PDF
Package pdftools has a powerful function, called pdf_data, to import data from a PDF. This function returns a list of data.frame, one per pages in the file, with one row for each text box and columns corresponding to sizes, coordinates and values of these text boxes.
library(pdftools)
metapdf <- pdf_data("2019-04-03-pdf-vs-html_files/test_form.pdf")
head(metapdf[[1]])## # A tibble: 6 x 6
## width height x y space text
## <int> <int> <int> <int> <lgl> <chr>
## 1 49 11 0 0 FALSE Introduction
## 2 4 11 0 781 TRUE 1
## 3 12 11 7 781 TRUE sur
## 4 4 11 22 781 FALSE 2
## 5 287 11 323 0 FALSE https://docs.google.com/forms/d/10UqbE6edL7K6G~
## 6 100 19 77 80 FALSE IntroductionThis function was perfect as the returned format was nearly the same as the one I wanted. To really obtained the final format, I just bound all data frames by row being careful of keeping a pages identifier.
library(pdftools)
metapdf <- bind_rows(metapdf, .id = "page")And that’s it ! With only one function used once, all the wanted data were extracted from the PDF. The rest is “just” data wrangling. At this point, I was convinced that the work was nearly finished but tidying the data frame meant finding some rules to get the finale data frame from the ones returned by the function. And finding these rules was not as obvious as I thought.
Tidying
Putting lines together
I selected all text boxes with a height equal to 10 as it seemed to be the height for question titles and answer choices. I then decided to reduce the data frame from one row per text box to one row per line in the PDF. The what-I-believed-being-an-acceptable-assumption was that all text boxes were in a same line if, in a page, they share the same y-coordinate. I collapsed text boxes sharing the same y-coordinate in a new column called lines and removed duplicated row based on this new column.
metapdf <- metapdf %>%
filter(height == 10) %>%
group_by(page, y) %>%
mutate(lines = str_c(text, collapse = " ")) %>%
distinct(lines, .keep_all = TRUE) %>%
ungroup()Identifying sentences
The second step was to identifying “sentences” among lines. The idea here was that a sentence starts either by one or two digits following by a dot and a space (question titles) or by an uppercase letter following by a lowercase letter or a space (answer choices). If a question title or an answer choice spreads on several lines of the PDF, I wanted to be able to consider all lines in the same sentence. I created a sentences_group column by a) detecting each text box starting a sentence using regular expressions, b) applying a cumulative sum on the boolean so each line belonging to the same sentence have the same sum value and c) transforming to string by adding a prefix. This column helped me collapsing sentences and removing duplicated ones.
metapdf <- metapdf %>%
mutate(sentences_group = str_detect(lines,"(^\\d{1,2}\\.\\s|^[:upper:]([:lower:]|\\s))") %>%
cumsum() %>%
paste0("sent", .)) %>%
group_by(sentences_group) %>%
mutate(sentences = str_c(lines, collapse = " ")) %>%
distinct(sentences_group, .keep_all = TRUE) %>%
ungroup()
select(metapdf, lines, sentences_group, sentences)
## # A tibble: 44 x 3
## lines sentences_group sentences
## <chr> <chr> <chr>
## 1 Event Timing: January 4th-~ sent1 Event Timing: January 4th-6th, 2~
## 2 Event Address: 123 Your St~ sent2 Event Address: 123 Your Street Y~
## 3 Contact us at (123) 456-78~ sent3 Contact us at (123) 456-7890 or ~
## 4 1. Name * sent4 1. Name *
## 5 2. Email * sent5 2. Email *
## 6 3. Organization * sent6 3. Organization *
## 7 4. What days will you atte~ sent7 4. What days will you attend? *
## 8 Check all that apply. sent8 Check all that apply.
## 9 Day 1 sent9 Day 1
## 10 Day 2 sent10 Day 2
## # ... with 34 more rowsDistinguishing question blocs
The method used to create column sentences_group was used to create column question_bloc which identifies the lines that belong to the same question i.e. the question titles and the answer choices. Value bloc0 corresponded to text before the first question so I removed it.
metapdf <- metapdf %>%
mutate(question_bloc = paste0("bloc", cumsum(str_detect(lines, "^\\d{1,2}\\.\\s")))) %>%
filter(question_bloc != "bloc0")Getting question types
For each question bloc, I was able to get the questions types thanks to the default behavior of Google Forms adding a sentence after each question title to specify how many answers can be given. These sentences were then removed and an identifier was given for sentences in each question bloc.
metapdf <- metapdf %>%
group_by(question_bloc) %>%
mutate(types = case_when("Check all that apply." %in% sentences ~ "multiple",
"Mark only one oval." %in% sentences ~ "unique",
"Mark only one oval per row." %in% sentences ~ "grid",
length(sentences) == 1 ~ "free")) %>%
filter(!is.element(sentences, c("Check all that apply.", "Mark only one oval.",
"Mark only one oval per row."))) %>%
mutate(id = 1:n()) Isolating questions and answer choices
The previously created identifier (id) enables the differenciation between question titles (id == 1) and answer choices (id != 1). However, grid needed a particular treatment as id == 2 corresponded to columns names and others ids corresponded to rows names. I first isolated this values, then created the choices and questions columns to finally put back answer choices values for grid questions.
# Grid question
columns <- metapdf %>%
filter(types == "grid" & id == 2) %>%
pull(sentences) %>%
str_split("\\s") %>%
unlist()
rows <- metapdf %>%
filter(types == "grid" & !id %in% 1:2) %>%
pull(sentences)
# Creating questions and choices columns
metapdf <- metapdf %>%
mutate(choices = list(sentences[-1])) %>%
ungroup %>%
filter(id == 1) %>%
select(questions = sentences, types, choices)
# Adding choices values for grid question
metapdf$choices[metapdf$types == "grid"] <- list(c(row = rows, column = columns))Cleaning
The last step was to do some cleaning. I removed the * that was present at the end of question titles where an answer was required.
metapdf <- metapdf %>% mutate(questions = str_replace(questions, "\\*", "") %>% str_trim)
metapdf
## # A tibble: 9 x 3
## questions types choices
## <chr> <chr> <list>
## 1 1. Name free <chr [0]>
## 2 2. Email free <chr [0]>
## 3 3. Organization free <chr [0]>
## 4 4. What days will you attend? multiple <chr [3]>
## 5 5. Dietary restrictions unique <chr [6]>
## 6 6. I understand that I will have to pay $$ upon arrival multiple <chr [1]>
## 7 7. What will you be bringing? multiple <chr [5]>
## 8 8. How did you hear about this event? unique <chr [5]>
## 9 9. What times are you available? grid <chr [9]>A try with rvest
If you have never heard about HTML and CSS, maybe what comes after will be a bit obscure but I think this is unlikely. Content of a web page is stored in HTML page and webscraping is collecting these data. This is why the rvest package was made for and I obviously used it for this task
rvest, selectorGadget and CSS selector
Basically, a HTML page has a tree structure (called a DOM for Document Object Model) where each node contains objects. Function read_html makes possible to read and import this DOM into R.
library(rvest)
form <- read_html("2019-04-03-pdf-vs-html_files/test_form.html")
form
form %>% html_nodes("body") %>% html_children() %>% length()## {html_document}
## <html lang="en" class="freebird">
## [1] <head>\n<meta http-equiv="Content-Type" content="text/html; charset=UTF-8 ...
## [2] <body dir="ltr" class="freebirdLightBackground isConfigReady" id="wizView ...
## [1] 23The first node of our page is called html and contains two children nodes called respectively head and body. If we take a look at the children nodes of body, we see that there are 23 of them. These nodes can have children too and so on.
Structure of a node looks like <name attr1 = "value1" attr2 = "value2"> object </name> where <name>...</name> is a tag (with a specific name) which can have several attributes (attr1, attr2) with specific values ("value1, "value2"). Tags delimit objects that can be other nodes or text content of the HTML page.
Nodes attributes can be CSS selectors. These are used to link nodes to CSS stylesheet which defined how content of the node will be displayed on a web browser. Name of these attributes are either id or class. CSS selectors are like identifiers of nodes and this is how they are used in webscraping by selecting nodes with specific id or class.
The real challenge here is to use the right CSS selectors. This is where SelectorGadget steps in. As Hadley Wickham wrote in this vignette of rvest package, “Selectorgadget is a javascript bookmarklet that allows you to interactively figure out what css selector you need to extract desired components from a page.” SelectorGadget is easy to use and the corresponding vignette makes clear how to use it so I won’t explain it in details in this post.
Scraping the data
Question blocs
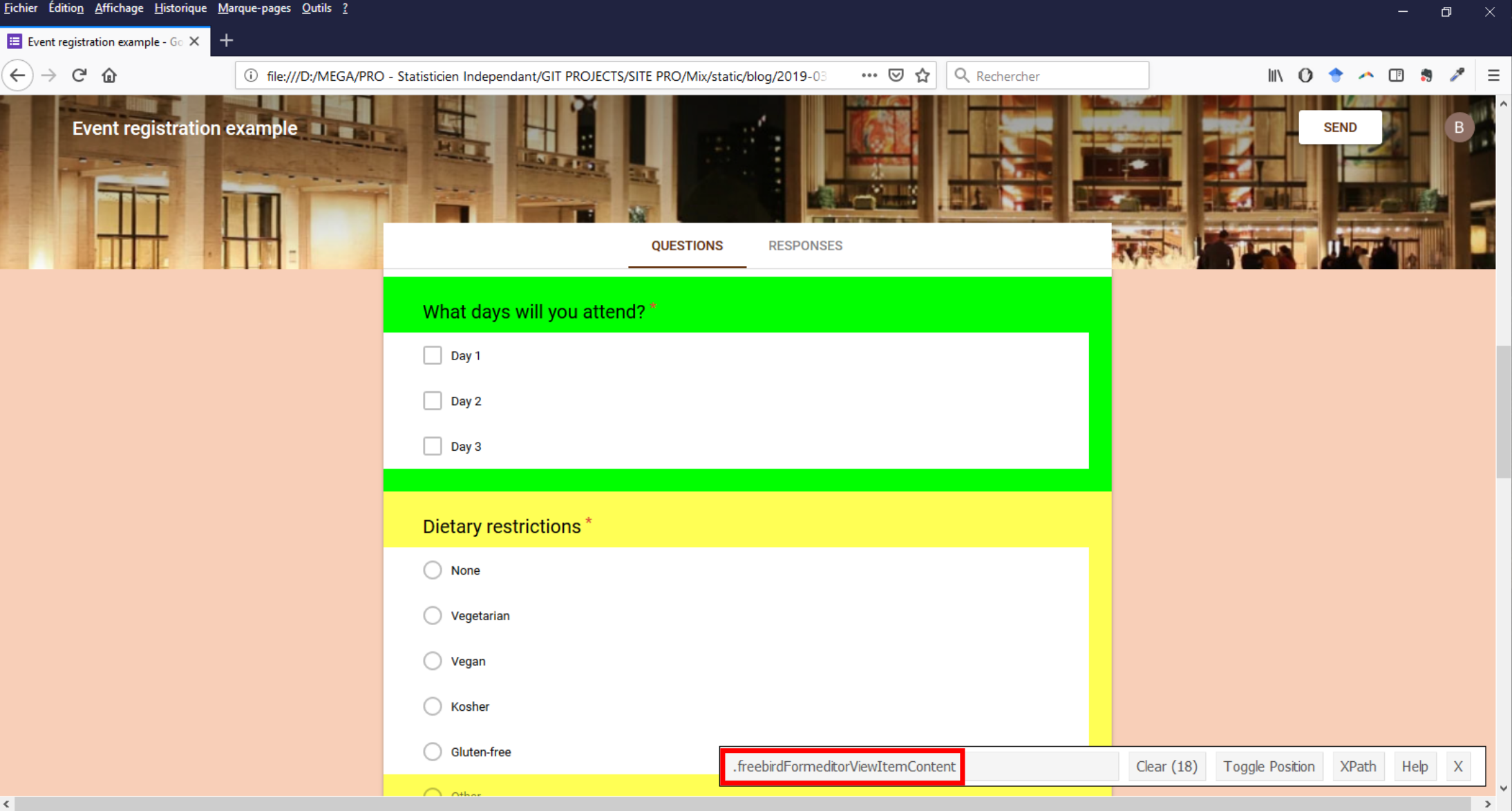
Question blocs (question titles + answer choices) were contained in nodes with freebirdFormeditorViewItemContent value for CSS class.
In this image (click to enlarge), GadgetSelector tells us that there are 18 nodes with these class value when there are only 9 questions in the questionnaire. I wasn’t able to precisely identify what were the extra 9 but I was able to determine how selecting the 9 ones of interest : they all contained a node with CSS class equal to freebirdFormeditorViewItemMinimizedTitleRow that itself contained a text corresponding to the question title.
prefix <- function(.x) {paste0(".freebirdFormeditorView", .x)}
question_blocs <- form %>%
html_nodes(prefix("ItemContent")) %>%
keep(~html_nodes(.x, prefix("ItemMinimizedTitleRow")) %>%
length() != 0)Question titles
As previously said, question titles were stored in nodes with class value equal to freebirdFormeditorViewItemMinimizedTitleRow.
questions <- question_blocs %>%
html_nodes(prefix("ItemMinimizedTitleRow")) %>%
html_text()
questions
## [1] "Name*"
## [2] "Email*"
## [3] "Organization*"
## [4] "What days will you attend?*"
## [5] "Dietary restrictions*"
## [6] "I understand that I will have to pay $$ upon arrival*"
## [7] "What will you be bringing?*"
## [8] "How did you hear about this event?*"
## [9] "What times are you available?*"Answer choices and question types
The next step was to get answer choices and question types by applying a function on each question bloc. This function contained if..else... statements depending on the type of questions.
Both kind of multiple choices questions were in nodes with class freebirdFormeditorViewQuestionBodyChoicelistbodyOmniList class. Answer choices were stored in attributes value in nodes with class exportInput and attribute aria-label equal to "option value". Some of these questions allowed an “Other” option as answer choice - this option required a special treatment as it was stored in different kind of nodes. Besides, question types between unique and multiple depended on data-list-type attribute value from the node with class freebirdFormeditorViewQuestionBodyChoicelistbodyOmniList.
Grid question were in question bloc that contains a node with class freebirdFormeditorViewQuestionBodyGridbodyRow. Text of nodes with only this class value corresponded to row names of the grid. Column names were stored as text in nodes with class freebirdFormeditorViewQuestionBodyGridbodyCell which were present in nodes with class freebirdFormeditorViewQuestionBodyGridbodyColumnHeader.
# To make code smaller
get_xpath <- function(.y) {
cl <- str_replace(prefix(.y), "\\.", "")
paste0('.//*[@class="', cl,'"]')
}
# To get choices and types
get_choices_and_types <- function(.nd) {
# Multiple choices questions
if (length(html_nodes(.nd, prefix("QuestionBodyChoicelistbodyOmniList"))) != 0) {
# Answer choices
choices <- html_nodes(.nd, ".exportInput") %>%
keep(~html_attr(.x, "aria-label") == "option value") %>%
html_attr(.,"value")
# For "Other" option
other <- html_nodes(.nd, prefix("OmnilistListPlaceholderOption")) %>%
html_attr(., "style") != "display:none"
if (other) choices <- c(choices, "Other:")
# Question type
dltype <- html_nodes(.nd, prefix("QuestionBodyChoicelistbodyOmniList")) %>%
html_attr("data-list-type")
if (dltype == "1") type <- "multiple" else type <- "unique"
# Grid question
} else if (length(html_nodes(.nd, prefix("QuestionBodyGridbodyRow"))) != 0) {
# Answer choices
choices <- c(
row = html_nodes(.nd, xpath = get_xpath("QuestionBodyGridbodyRow")) %>%
html_text(),
column = html_nodes(.nd, prefix("QuestionBodyGridbodyColumnHeader")) %>%
html_nodes(xpath = get_xpath("QuestionBodyGridbodyCell")) %>%
html_text()
)
# Question type
type <- "grid"
# Free question
} else {
choices <- character(0)
type <- "free"
}
return(tibble(types = type, choices = list(choices)))
}
choices_types <- map_dfr(question_blocs, get_choices_and_types)Cleaning
Finally, the last step was to bind data of interest and apply the same cleaning as with the PDF.
metahtml <- bind_cols(questions = paste0(1:9, ". ", unlist(questions)), choices_types)
metahtml <- mutate(metahtml, questions = str_replace(questions, "\\*", "") %>% str_trim)
metahtml
## # A tibble: 9 x 3
## questions types choices
## <chr> <chr> <list>
## 1 1. Name free <chr [0]>
## 2 2. Email free <chr [0]>
## 3 3. Organization free <chr [0]>
## 4 4. What days will you attend? multiple <chr [3]>
## 5 5. Dietary restrictions unique <chr [6]>
## 6 6. I understand that I will have to pay $$ upon arrival multiple <chr [1]>
## 7 7. What will you be bringing? multiple <chr [5]>
## 8 8. How did you hear about this event? unique <chr [5]>
## 9 9. What times are you available? grid <chr [9]>Conclusion
We can first check if all columns in metapdf and metahtml are equals.
map2(metapdf, metahtml, all.equal)
## $questions
## [1] TRUE
##
## $types
## [1] TRUE
##
## $choices
## [1] TRUEWith two different kind of files, I was able to get the same result thanks to two amazing packages : pdftools and rvest. Scraping the HTML page could have been made with xml2 package as lots of rvest functions are wrappers around xml2 functions. The main difference is that xml2 functions only accept xpath expressions which from my point of view has a non easy syntax to master (but maybe I’m lazy). However, you can get xpath expressions from GadgetSelector which helps their utilisation along.
It was not the purpose of this work to tell if one way of extracting information is better than the other. I had fun with both and I was amazed by the power of both packages. There will be days when I’ll have to work with PDF, and I know that I can count on pdftools for that, as much as I am more than convinced by rvest to help me during my future webscraping.
If I had one difference to point out is that rules to wrangle data from the PDF strongly depend on how the PDF file is structured. The large diversity of possible structure probably makes difficult to define a reproducible strategy while for HTML page strategy is always the same : find value of CSS selectors for the data you want with SelectorGadget and apply the appropriate function to extract it. In the end, it might not be faster but it sure makes the workflow more obvious.
I hope you enjoyed reading this first post as much as I enjoyed writing it. Don’t be shy ! Leave a comment for anything and let me know if you would have handled things differently.